Selected publications (a full list also available)
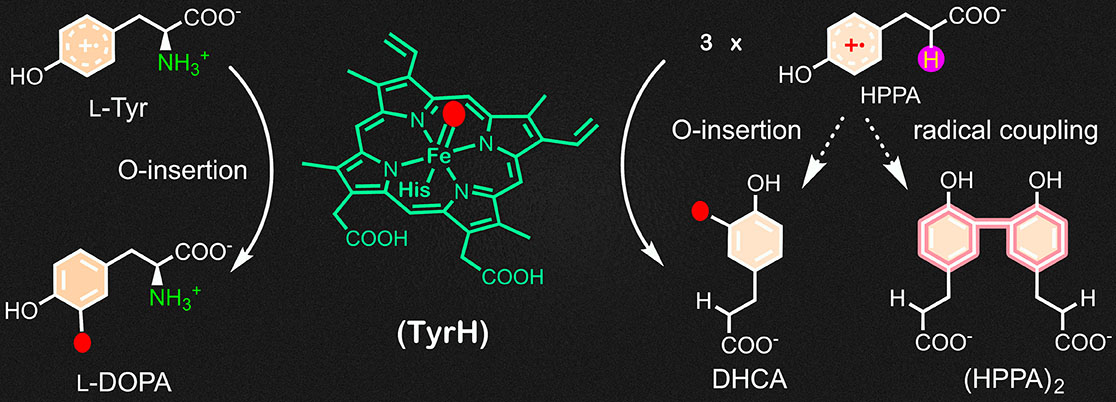
Substrate analogs implicate a free radical pathway in tyrosine hydroxylase catalysis
Traore ES, Wang Y, Griffith WP, and Liu A
ACS Catalysis, 2025, 15, 18270-18281 (DOI: 10.1021/acscatal.5c05776)
Narrator: Ephrahime Traore (pending)
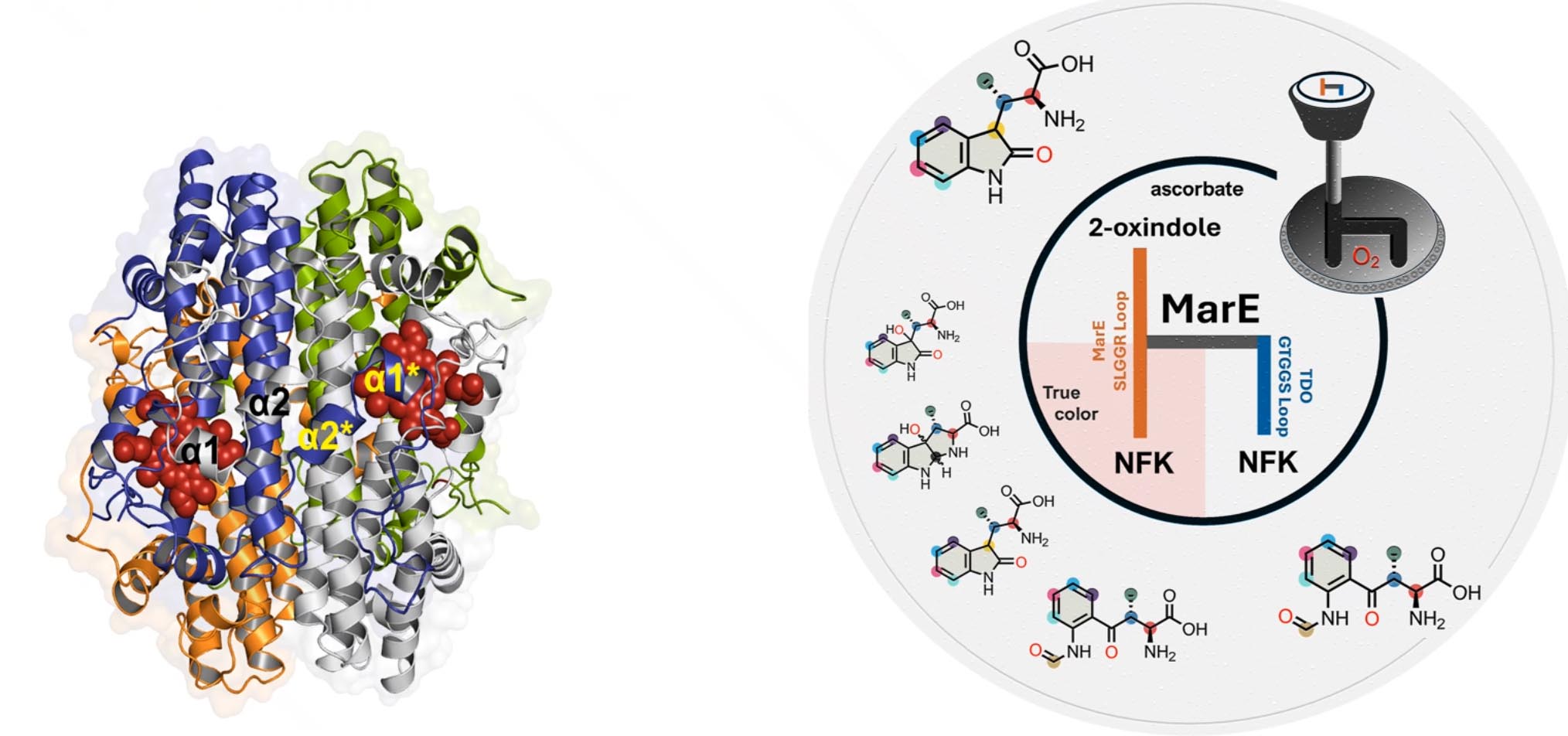
Deciphering tryptophan oxygenation: Key modulators of 2-oxindole formation in MarE
Nguyen R, Shin, I, and Liu A
Angew. Chem. Int. Ed., 2025, 64(35), e202510848 (DOI: 10.1002/anie.202510848)
Narrator: Romie Nguyen
Protein-derived cofactors: Chemical innovations expanding enzyme catalysis
Graciano A and Liu A
Chem. Soc. Rev., 2025, 54(9) 4502 - 4530 (DOI: 10.1039/d4cs00981a)
Catalase-peroxidase (KatG): A potential frontier in tuberculosis drug development
Liu A
Crit. Rev. Biochem. Mol. Biol., 2024, 59(6), 434-446 (DOI: 10.1080/10409238.2025.2470630)
Structural insights into 2-oxindole-forming monooxygenase MarE: Divergent architecture and substrate positioning versus tryptophan dioxygenases
Shin I, Nguyen RC, Montoya SR, and Liu A*
J. Biol. Chem., 2025, 301(3), 108241 (1-14) (DOI: 10.1016/j.jbc.2025.108241)
α-Amino-β-carboxymuconate-ε-semialdehyde decarboxylase catalyzes enol/keto tautomerization of oxaloacetate
Yang Y*, Davis I, Altman RA, and Liu A*
J. Biol. Chem., 2024, 300(11), 107878 (1-11) (DOI: 10.1016/j.jbc.2024.107878)
Indole N-linked hydroperoxyl adduct of protein-derived cofactor modulating catalase-peroxidase functions
Li J, Duan R, Traore ES, Davis I, Nguyen RC, Griffith WP, Goodwin DC, Jarzecki AA, and Liu A*
Angew. Chem. Int. Ed., 2024, 63(49), e202407018 (1-10) (DOI: 10.1002/anie.202407018)
Cobalt(II)-substituted cysteamine dioxygenase oxygenation proceeds through a cobalt(III)-superoxo complex
Li J*, Duan R, and Liu A*
J. Am. Chem. Soc., 2024, 146(27), 18292-18297 (DOI: 10.1021/jacs.4c01871)

In situ structural observation of a substrate- and peroxide-bound high-spin
ferric-hydroperoxo intermediate in P450 enzyme CYP121
Nguyen RC, Davis I, Dasgupta M, Wang Y, Simon P,
Butryn A, Makita H, Bogacz I, Dornevil K, Aller P, Bhowmick A, Chatterjee R,
Kim I-S, Zhou T, Mendez D, Paley, D, Fuller F, Alonso Mori R, Batyuk A,
Sauter N, Brewster A, Orville AM, Yachandra V, Yano J, Kern J,* and Liu A*
J. Am. Chem. Soc., 2023, 145(46), 25120-25133 (DOI: 10.1021/jacs.3c04991)

Charge maintenance during catalysis in non-heme iron oxygenases
Traore ES and Liu A*
ACS Catalysis, 2022, 12(10), 6191-6208 (DOI: 10.1021/acscatal.1c04770)

A new regime of heme-dependent aromatic oxygenase superfamily
Shin I, Wang Y, and Liu A*
![]() Proc. Natl. Acad. Sci. U.S.A., 2021,
118(43), e210656 (1-10) (DOI: 10.1073/pnas.2106561118)
Proc. Natl. Acad. Sci. U.S.A., 2021,
118(43), e210656 (1-10) (DOI: 10.1073/pnas.2106561118)

Crystal
structure of human cysteamine dioxygenase provides a structural rationale for
its function as an oxygen sensor
Wang Y, Shin I, Li J, and Liu A*
J. Biol.
Chem., 2021, 297(4), 101176 (1-10) (DOI: 10.1016/j.jbc.2021.101176)

![]()
Narrator: Yifan (Amber) Wang
Molecular
rationale for partitioning between C-H and C-F bond activation in
heme-dependent tyrosine hydroxylase
Wang Y, Davis I, Shin I, Xu H, and Liu A*
J. Am.
Chem. Soc., 2021, 143(12), 4680-4693 (DOI: 10.1021/jacs.1c00175)

![]()
Narrator: Yifan (Amber) Wang
MP4, 8'52"
A novel
catalytic heme cofactor in SfmD with a single
thioether bond and a bis-His ligand set revealed by de novo crystal
structural and spectroscopic study
Shin I, Davis I, Nieves-Merced K, Wang Y, McHardy
S, and Liu A*
Chemical
Science, 2021, 12(11), 3984-3998 (DOI: 10.1039/D0SC06369J)

![]()
Inchul Shin
(MP4,
13'24")
Diflunisal derivatives as modulators of ACMS decarboxylase targeting the tryptophan-kynurenine pathway
Yang Y, Borel T, de Azambuja F, Johnson D, Sorrentino JP, Udokwu C, Davis I, Liu A*, and Altman RA*
J. Med. Chem., 2021, 64(1), 797-811
(DOI: 10.1021/acs.jmedchem.0c01762)

Heme binding to HupZ with a C-terminal tag from Group A Streptococcus
Traore ES, Li J, Chiura T, Geng J, Sachla A, Yoshimoto F, Eichenbaum Z, Davis I, Max P*, and Liu A*
Molecules, 2021, 26(3), 549 (1-19)
(DOI: 10.3390/molecules26030549)

![]()
Narrator: Ephrahime S. Traore
(LiveSlides: pending)
Formation of monofluorinated radical cofactor in galactose oxidase through copper-mediated
C−F bond scission
Li J, Davis I, Griffith WP, and Liu A*
J. Am. Chem. Soc., 2020, 142(44), 18753-18757 (DOI: 10.1021/jacs.0c08992)

![]()
Narrator: Jiasong Li
(MP4,
7'39")
Observing
3-hydroxyanthranilate-3,4-dioxygenase in action through a crystalline lens
Wang Y, Liu KF, Yang Y, Davis I, and Liu A*
![]() Proc. Natl. Acad. Sci. U.S.A., 2020,
117(33) 19720-19730 (PNAS Direct Submission) (DOI: 10.1073/pnas.2005327117)
Proc. Natl. Acad. Sci. U.S.A., 2020,
117(33) 19720-19730 (PNAS Direct Submission) (DOI: 10.1073/pnas.2005327117)

![]()
Narrators: Yifan Wang & Ian Davis
(an accompany
enzyme action movie published by PNAS)
Wang Y, Davis I, Yang Y, Chen Y, Naik SG, Griffith WP, and Liu A*
J. Biol. Chem., 2020, 295(33), 11789-11802 (DOI: 10.1074/jbc.RA120.013915)

![]()
Narrator: Yifan Wang
Kinetic and
spectroscopic characterization of the catalytic ternary complex of tryptophan
2,3-dioxygenase
Geng J, Weitz AC, Dornevil K, Hendrich MP,
and Liu A*
Biochemistry, 2020,
59(30), 2813-2822 (DOI: 10.1021/acs.biochem.0c00179)

Carbon-fluorine
bond cleavage mediated by metalloenzymes
Wang Y and Liu A*
Chem. Soc. Rev., 2020, 49(14), 4906-4925 (DOI: 10.1039/C9CS00740G)

Substrate-assisted hydroxylation and O-demethylation in the peroxidase-like cytochrome P450 enzyme CYP121
Nguyen RC, Yang Y, Wang Y, Davis I, and Liu A*
ACS Catalysis, 2020, 10(2), 1628-1639 (DOI: 10.1021/acscatal.9b04596)

![]()
Narrator: Romie C. Nguyen
Crystal structures of L-DOPA dioxygenase from Streptomyces sclerotialus
Wang Y, Shin I, Fu Y, Colabroy KL*, and Liu A*
Biochemistry, 2019,
58(52), 5339-5350 (DOI: 10.1021/acs.biochem.9b00396)
(invited contribution for a special issue of Current Topics in Mechanistic Enzymology 2019)

![]()
Narrators:
Yifan Wang & Inchul Shin
Quaternary structure of α-amino-β-carboxymuconate-ε-semialdehyde
decarboxylase (ACMSD) controls its activity
Yang Y, Davis I, Matsui T, Rubalcava I, and Liu A*
J. Biol. Chem., 2019, 294(30), 11609-11621 (DOI: 10.1074/jbc.RA119.009035) (LiveSlide video presentation) featured as a JBC cover story
Narrator: Yu Yang
Biocatalytic carbon−hydrogen and carbon−fluorine bond cleavage through
hydroxylation promoted by a histidyl-ligated heme enzyme
Wang Y, Davis I, Shin I, Wherritt DJ, Griffith WP,
Dornevil K, Colabroy KL, and Liu A*
ACS
Catalysis, 2019, 9(6), 4764-4776 (DOI: 10.1021/acscatal.9b00231)
featured as an ACS Editors Choice article

Narrators:
Yifan Wang and Ian Davis
(Short version published by ACS: 8 slides)
Probing the Cys-Tyr cofactor biogenesis in cysteine dioxygenase by the
genetic incorporation of fluorotyrosine
Li J, Koto, T, Davis I, and Liu A*
Biochemistry, 2019,
58(17), 2218-2227 (DOI: 10.1021/acs.biochem.9b00006)
(a highly cited original paper in the journal)

Cleavage of a carbon−fluorine bond by an engineered cysteine dioxygenase
Li J, Griffith WP, Davis I, Shin I, Wang J, Li F,
Wang Y, Wherritt D, and Liu A*
Nature
Chemical Biology, 2018, 14(9), 853-860 (DOI: 10.1038/s41589-018-0085-5)

Adapting to oxygen: 3-hydroxyanthranilate 3,4-dioxygenase employs loop
dynamics to accommodate two substrates with disparate polarities
Yang Y, Liu F*, and Liu A*
J. Biol.
Chem., 2018, 293(27), 293, 10415-10424 (DOI: 10.1074/jbc.RA118.002698) (featured as a JBC cover story)

Cofactor biogenesis in cysteamine dioxygenase: C−F bond cleavage with
genetically incorporated unnatural tyrosine
Wang Y, Griffith WP, Li J, Koto T, Wherritt D,
Fritz E, and Liu A*
Angew. Chem. Int. Ed., 2018,
57(27), 8149-8153 (DOI: 10.1002/anie.201803907)

Reassignment of the human aldehyde dehydrogenase ALDH8A1 (ALDH12) to the
kynurenine pathway in tryptophan catabolism
Davis I, Yang Y, Wherritt D, and Liu A*
J. Biol.
Chem., 2018, 293(25), 9594-9603 (DOI: 10.1074/jbc.RA118.003320)

Stepwise O-atom transfer in heme-based tryptophan dioxygenase: Role of
substrate ammonium in epoxide ring opening
Shin I, Ambler BR, Wherritt DJ, Griffith WP,
Maldonado AC, Altman RA, and Liu A*
J. Am.
Chem. Soc., 2018, 140(12), 4372-4379 (DOI: 10.1021/jacs.8b00262)

High-frequency/high-field EPR and theoretical studies of tryptophan-based
radicals
Davis I, Koto T, Terrell JR, Kozhanov A, Krzystek
J, and Liu A*
J. Phys.
Chem. A, 2018, 122(12), 3170-3176 (DOI: 10.1021/acs.jpca.7b12434)

Backbone dehydrogenation in pyrrole-based pincer ligands
Krishnan VM, Davis I, Baker TM, Curran DJ, Arman H, Neidig ML, Liu A, and Tonzetich ZJ*
Inorg. Chem., 2018, 57(15), 9544-9553 (DOI: 10.1021/acs.inorgchem.8b01643)

Radical trapping study of the relaxation of bis-Fe(IV) MauG
Davis I, Koto T, and Liu A*
Reactive
Oxygen Species, 2018, 5(13), 46-55 (DOI: 10.20455/ros.2018.801)

Probing ligand exchange in the P450 enzyme CYP121 from Mycobacterium tuberculosis:
Dynamic equilibrium of the distal heme ligand as a function of pH and temperature
Fielding AJ, Dornevil K, Ma L, Davis I, and Liu A*
J. Am. Chem. Soc., 2017, 139(48), 17484-17499 (DOI: 10.1021/jacs.7b08911)

Cross-linking of dicyclotyrosine by the cytochrome P450 enzyme CYP121 from Mycobacterium tuberculosis proceeds through a catalytic shunt pathway
Dornevil K, Davis I, Fielding AJ, Terrell JR, Ma L, and Liu A*
J. Biol. Chem., 2017, 292(33), 13645-13657 (DOI: 10.1074/jbc.M117.794099)

Hypertryptophanemia due to tryptophan
2,3-dioxygenase deficiency
Ferreira F,* Shin I, Sosova I, Dornevil K, Jain Shailly, Dewey D, Liu F, and Liu A*
Mol.
Genet. Metab., 2017, 120(4), 317-324
(DOI: 10.1016/j.ymgme.2017.02.009)

Mutual synergy between catalase and peroxidase activities of the bifunctional enzyme
KatG is facilitated by electron-hole hopping within the enzyme
Njuma OJ, Davis I, Ndontsa EN, Krewall JR, Liu A, and Goodwin DC*
J. Biol. Chem., 2017, 292(45), 18408-18421
(DOI: 10.1074/jbc.M117.791202)

Oxygen activation by mononuclear nonheme iron dioxygenases involved in the degradation of aromatics
Wang Y, Li J, and Liu A*
J. Biol.
Inorg. Chem., 2017, 22(2), 395-405 (DOI: 10.1007/s00775-017-1436-5)
(invited review article for a special issue of the journal under the theme of 60
Years of Oxygen Activation)

A pitcher-and-catcher mechanism drives endogenous substrate isomerization by
dehydrogenase in kynurenine metabolism
Yang Y, Davis I, Ha U, Wang Y, Shin I, and Liu
A*
J. Biol.
Chem., 2016, 291(51), 26252-26261 (DOI: 10.1074/jbc.M116.759712)
(featured as "Papers of the
Week and selected, after publication, into a collection of Enzymology
virtual issue)

Control of carotenoid biosynthesis through a heme-based cis-trans isomerase
Beltran J, Kloss B, Hosler JP, Geng J, Liu A, Modi A, Dawson JH, Sono M, Shumskaya M, Ampomah-Dwamena C, Love JD, and Wurtzel ET*
Nature
Chemical Biology, 2015, 11(8), 598-605 (DOI: 10.1038/nchembio.1840)

What is the
tryptophan kynurenine pathway and why is it important to neurotherapeutics? (Invited
Editorial)
Davis I and Liu A*
Expert
Review of Neurotherapeutics, 2015, 15(7), 719-721 (DOI: 10.1586/14737175.2015.1049999)

An iron reservoir to the catalytic metal: The rubredoxin iron in an extradiol dioxygenase
Liu F, Geng J, Gumpper RH, Barman A, Davis I,
Ozarowski A, Hamelberg D, and Liu A*
J. Biol.
Chem., 2015, 290(25), 15621-15634 (DOI: 10.1074/jbc.M115.650259)

Probing bis-Fe(IV) MauG:
Experimental evidence for the long-range charge-resonance model
Geng J, Davis I, and Liu A*
Angew. Chem. Int. Ed., 2015,
54, 3692-3696 (DOI: 10.1002/anie.201410247)

Crystallographic and spectroscopic snapshots reveal a dehydrogenase in
action
Huo L, Davis I, Liu F, Andi B, Esaki S, Hiroaki I,
Li T, Hasegawa Y, Orville AM, and Liu A*
Nature
Communication, 2015, 6:5935 (1-10) (DOI: 10.1038/ncomms6935)
(defining the structure of a kynurenine pathway dehydrogenase and sp3-to-sp2 transition
intermediates during catalysis)

Human α-amino-β-carboxymuconate-ε-semialdehyde
decarboxylase (ACMSD): A structural and mechanistic unveiling
Huo L, Liu F, Hiroaki I, Li T, Hasegawa Y,
and Liu A*
Proteins, 2015,
83(1), 178-187 (DOI: 10.1002/prot.24722)

Bis-Fe(IV): Nature's sniper for long-range
oxidation
Geng J, Davis I, Liu F, and Liu A*
J. Biol. Inorg. Chem., 2014, 19(7), 1057-1067 (Invited Article) (DOI: 10.1007/s00775-014-1123-8)
Amidohydrolase
Superfamily
Liu A* and Huo L
Encyclopedia of Life Sciences, 2014, 1-11 (DOI: 10.1002/9780470015902.a0020546.pub2)
Power of two: Arginine 51 and arginine 239* from a neighboring subunit are
essential for catalysis in α-amino-β-carboxymuconate-ε-semialdehyde
decarboxylase
Huo L, Davis I, Chen L, and Liu A*
J. Biol.
Chem., 2013, 288(43), 30862-30871 (DOI: 10.1074/jbc.M113.496869)

Pirin is an iron-dependent redox regulator
of NF-κB
Liu F, Rehmani I, Esaki S, Fu R, Chen L, Serroano
V, and Liu A*
![]() Proc.
Natl. Acad. Sci. U.S.A., 2013, 110(24), 9722-9727 (PNAS Direct
Submission) (DOI: 10.1073/pnas.1221743110)
Proc.
Natl. Acad. Sci. U.S.A., 2013, 110(24), 9722-9727 (PNAS Direct
Submission) (DOI: 10.1073/pnas.1221743110)

(**
Faculty of 1000 recommended article )
Tryptophan-mediated charge-resonance stabilization in the bis-Fe(IV) redox state of MauG
Geng J, Dornevil K, Davidson VL, and Liu A*
![]() Proc.
Natl. Acad. Sci. U.S.A., 2013, 110(24), 9639-9644 (PNAS Direct
Submission) (DOI: 10.1073/pnas.1301544110)
Proc.
Natl. Acad. Sci. U.S.A., 2013, 110(24), 9639-9644 (PNAS Direct
Submission) (DOI: 10.1073/pnas.1301544110)

Diradical intermediate within the context of tryptophan tryptophylquinone
biosynthesis
Yukl ET, Liu F, Krzystek J, Shin S, Jensen LMR,
Davidson VL, Wilmot CM,* and Liu A*
![]() Proc. Natl. Acad.
Sci. U. S. A., 2013, 110(12), 4569-4573 (PNAS Direct Submission) (DOI: 10.1073/pnas.1215011110)
Proc. Natl. Acad.
Sci. U. S. A., 2013, 110(12), 4569-4573 (PNAS Direct Submission) (DOI: 10.1073/pnas.1215011110)

* Faculty of 1000 recommended article
An unexpected copper catalyzed 'reduction' of an arylazide
to amine through the formation of a nitrene intermediate
Peng H, Dornevil K, Draganov A, Chen W, Dai C,
Nelson WH, Liu A,* and Wang B*
Tetrahedron,
2013, 69, 5079-5085 (dedicated to the memory of Professor William H.
Nelson) (DOI: 10.1016/j.tet.2013.04.091)

Chemical rescue of the distal histidine mutants of tryptophan
2,3-dioxygenase
Geng J, Dornevil K, and Liu A*
J. Am. Chem. Soc., 2012, 134, 12209-12218 (DOI: 10.1021/ja304164b)
Evidence for a dual role of an active site histidine in α -amino-β - carboxymuconate- ε - semialdehyde decarboxylase
Huo L, Fielding AJ, Chen Y, Li T, Iwaki H, Hosler JP, Chen L, Hasegawa Y, Que Jr, L, and Liu A*
Biochemistry,
2012, 51(29), 5811-5821 (DOI: doi.org/10.1021/bi300635b)

The role of calcium in metalloenzyme: Effects of calcium removal on the
axial ligation geometry and magnetic properties of the catalytic diheme center in MauG
Chen Y, Naik SG, Krzystek J, Shin S, Nelson WH,
Xue S, Yang JJ, Davidson VL, and Liu A*
Biochemistry, 2012, 51,
1586-1597 (DOI: 10.1021/bi201575f)

The reactivation mechanism of tryptophan 2,3-dioxygenase by hydrogen
peroxide
Fu R, Gupta R, Geng J, Dornevil K, Wang S, Hendrich MP,
and Liu A*
J. Biol.
Chem., 2011, 286(30), 26541-26554 (DOI: 10.1074/jbc.M111.253237)

Mutagenesis
of tryptophan199 suggests that electron hopping is required for MauG-dependent tryptophan tryptophylquinone
biosynthesis
Tarboush NA, Jensen LMR, Yukl ET, Geng J, Liu A, Wilmot CM,
and Davidson VL
![]() Proc. Natl. Acad.
Sci. U. S. A., 2011, 108(41), 16956-16961 (PNAS Direct Submission) (DOI: 10.1073/pnas.1109423108)
Proc. Natl. Acad.
Sci. U. S. A., 2011, 108(41), 16956-16961 (PNAS Direct Submission) (DOI: 10.1073/pnas.1109423108)

EPR and
M�ssbauer spectroscopy show inequivalent hemes in tryptophan dioxygenase
Gupta R, Fu R,
Liu A, and Hendrich MP*
J. Am. Chem. Soc., 2010, 132(3), 1098-1109 (DOI: 10.1021/ja908851e)
Heme iron
nitrosyl complex of MauG reveals efficient redox
equilibrium between hemes with only one heme exclusively binding exogenous ligands
Fu R, Liu F, Davidson VL,
and Liu A*
Biochemistry,
2009, 48(49), 11603-11605 (DOI: 10.1021/bi9017544)

Electron Paramagnetic Resonance (EPR) in Enzymology
Liu A*
Wiley
Encyclopedia of Chemical Biology, 2008, 1,
591-601 (DOI: 10.1002/9780470048672.wecb668)
A catalytic
di-heme bis-Fe(IV) form of MauG, Alternative to an Fe(IV)=O porphyrin radical
Li X, Fu R, Lee
S, Krebs C, Davidson VL,* and Liu A*
![]() Proc.
Natl. Acad. Sci. U.S.A., 2008, 105(25), 8597-8600
(PNAS Direct Submission) (DOI: 10.1073/pnas.0801643105)
Proc.
Natl. Acad. Sci. U.S.A., 2008, 105(25), 8597-8600
(PNAS Direct Submission) (DOI: 10.1073/pnas.0801643105)

Kinetic and
physical evidence that the di-heme enzyme MauG
tightly binds to a biosynthetic precursor of methylamine dehydrogenase with
incompletely formed tryptophan tryptophylquinone
Li X, Fu R, Liu A*, and Davidson
VL*
Biochemistry, 2008, 47(9),
2908-2912 (DOI: 10.1021/bi702259w)

Amidohydrolase Superfamily
Liu A*, Li T, and Fu R
Encyclopedia
of Life Sciences, 2007, 1-8 (DOI: 10.1002/9780470015902.a0020546)
Detection of transient intermediates in the metal-dependent non-oxidative
decarboxylation catalyzed by α-amino-β-carboxymuconic-ε-semialdehyde
decarboxylase
Li T, Ma
J, Hosler JP, Davidson VL, and Liu A*
J. Am. Chem. Soc.,
2007, 129(30), 9278-9279 (DOI: 10.1021/ja073648le)

Crystallographic analysis of α-amino-β-carboxymuconic-ε-semialdehyde decarboxylase:
Insight into the active site and catalytic mechanism of a novel
decarboxylation reaction
Martynowski D., Eyobo Y.,
Li T, Yang K., Liu A,* and Zhang H*
Biochemistry, 2006, 45(35),
10412-10421 (DOI: 10.1021/bi060903q)

Transition
metal-catalyzed nonoxidative decarboxylation reactions
Liu A* and
Zhang H*
Biochemistry, 2006, 45(35),
10407-10411 (a New Concept paper) (DOI: 10.1021/bi061031v)

α-Amino-β-carboxymuconic-ε-semialdehyde decarboxylase (ACMSD)
is a new member of the amidohydrolase superfamily
Li T, Iwaki H,
Fu R, Hasegawa Y, Zhang H, Liu A*
Biochemistry, 2006, 45(21), 6628-6634 (DOI: 10.1021/bi060108c)
Kinetic and spectroscopic characterization of ACMSD from Pseudomonas
fluorescens reveals a pentacoordinate mononuclear metallocofactor
Li T, Walker AL, Iwaki H, Hasegawa Y, Liu A*
J. Am.
Chem. Soc., 2005, 127(35), 12282-12290 (DOI: 10.1021/ja0532234)
 ☺
☺