X-ray protein crystallography has been a new focus of our laboratory since the spring of 2011.
We have solved over 1,000 non-redundant structures and published 142 crystal structures and two small-angle X-ray scattering (SAXS) solution structures,
including some de novo protein structures and many reactive intermediate structures. Most of these structures are iron-based enzymes and a regulatory protein
of signal transductions. Overall, structural characterization has become an integrated efforts in our biochemical, spectroscopic and structural studies of
metallproteins/metalloenzymes and protein-based free radicals.
RCSB PDB database entries:
8TDQ, 8TDP, 8D5M, 8D0L, 8CZP, 8CZS, 7S5D, 7S0O, 7RJY, 7RJV, 7RJS, 7RJW, 7RJX, 7RUK,
7REI, 7KQR, 7KQS, 7KQT, 7KQU, 7KU8, 7KQS, 7KQR, 7KQT, 7KPZ, 7KQ2, 7K12, 7K13, 6XLR,
6XLT, 6XLS, 6VI5, 6VI6, 6VI7, 6VI8, 6VI9, 6VIA, 6VIB, 6VDP, 6VDQ, 6VDZ, 6VE0, 6UPG, 6UPI,
6NA9, 6NA8, 6NA7, 6NA2, 6NA1, 6NA0, 6N9Z, 6ON3, 6ON1, 6E87, 6EPR, 6N42, 6N43, 6BPR,
6BPX, 6BPS, 6BPT, 6BPU, 6BPV, 6BPW, 6CDH, 6CDN, 6BVP, 6BVQ, 6BVR, 6BVS, 6CD3, 6D60,
6D61, 6D62, 6MGS, 6MGT, 6BGF, 6BGM, 6C5P, 6C5O, 6BUK, 6BUJ, 6BUF, 6BUE, 6BUD, 6C0Q,
5W9U, 5W9V, 5W9T, 5WP2, 5V26, 5V27, 5V28, 5KJ5, 5KLK, 5KLL, 5KLM, 5KLN, 5KLO, 4R52,
4WZC, 4HSJ, 4L2N, 4NPI, 4OE2, 4OU2, 4OUB, 4OFC, 4I1W, 4I25, 4I26, 4I2R, 4IH3, 4IGM, 4IGN,
4I3P, 4HSL, 4HVO, 4HVQ, 4HVR, 4IFK, 4IFO, 4IFR, 4IG2, 4ERO, 4EWA, 4EWD, 4EWE, 4GUL,
4HLT, 4FB1, 4FA1, 4FA4, 4FA5, 4FA9, 4FAN, 4FAV, 4EPK, 4ERA, 4ERG, 4ERI, 2HBV, and 2HBX
Small-angle X-ray scattering (SAXS) solution structures authored:
SASDFN5 and SASDFM5
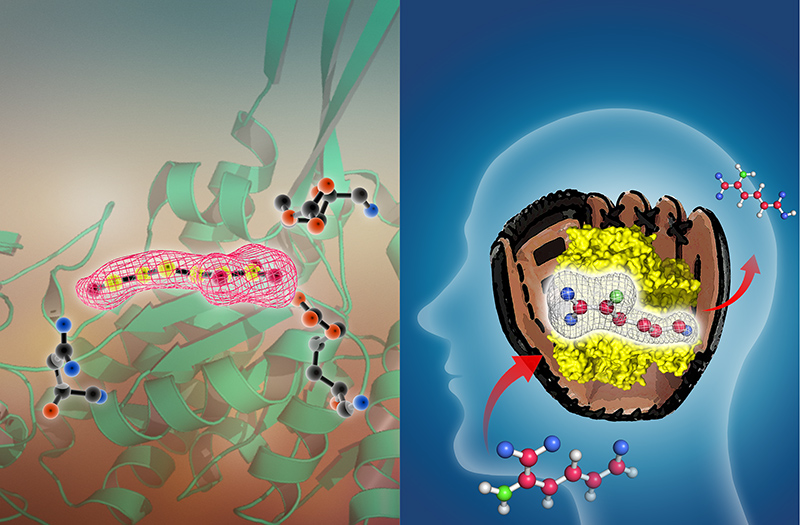
A pitcher-and-catcher mechanism drives endogenous substrate isomerization by a dehydrogenase in kynurenine metabolism
Yang Y, Davis I, Ha U, Wang Y, Shin I and Liu A*
J. Biol. Chem., 2016, 291(51), 26252-26261 (
doi:10.1074/jbc.M116.759712)
(This paper was designated by teh editors as a "Papers of the Week" and selected, after publication, in a collection of a representative snapshot of recent papers in the field of Enzymology in a
virtual issue).
Structures associated with this publication and released in the protein databank with the following PDB entries: 5KJ5, 5KLK, 5KLL, 5KLM, 5KLN, and 5KLO
Aldehyde dehydrogenases mediate the oxidation of highly reactive aldehyde compounds in the cell. This paper reveals a pitcher-and-catcher chemical mechanism of a hidden isomerization
performed by a kynurenine pathway aldehyde dehydrogenase prior to the expected redox reaction. The figure above shows a critical catalytic intermediate caught in action by protein X-ray crystallography.
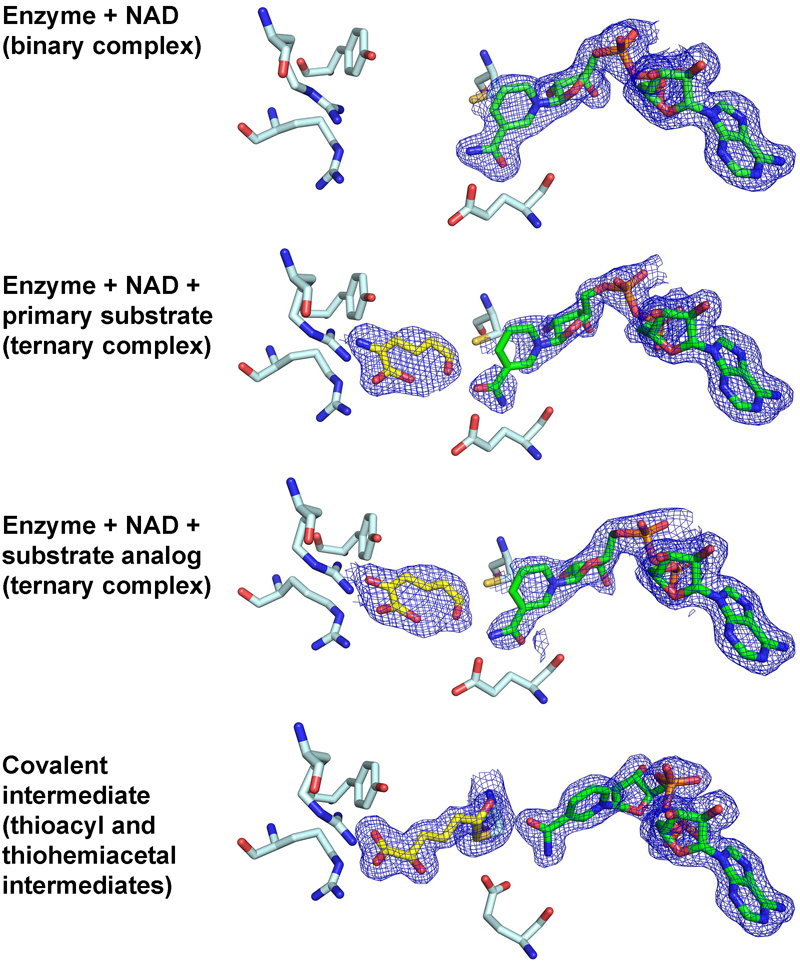
Capture of tetrahedral thiohemiacetal and thioacyl intermediates reveals an sp3-to-sp2 transition during an enzyme-mediated dehydrogenation
Huo L,^ Davis I,^ Liu F, Andi B, Esaki S, Hiroaki I, Li T, Hasegawa Y, Orville AM, and Liu A*
Nature Communications 2015, 6:5935 (DOI: 10.1038/ncomms6935).
Structures associated with this publication and released in the protein databank with following PDB entries: 4I25, 4I26, 4I1W, 4I2R, 4NPI, 4OE2, 4OU2, and 4OUB.
^: shared first authorship
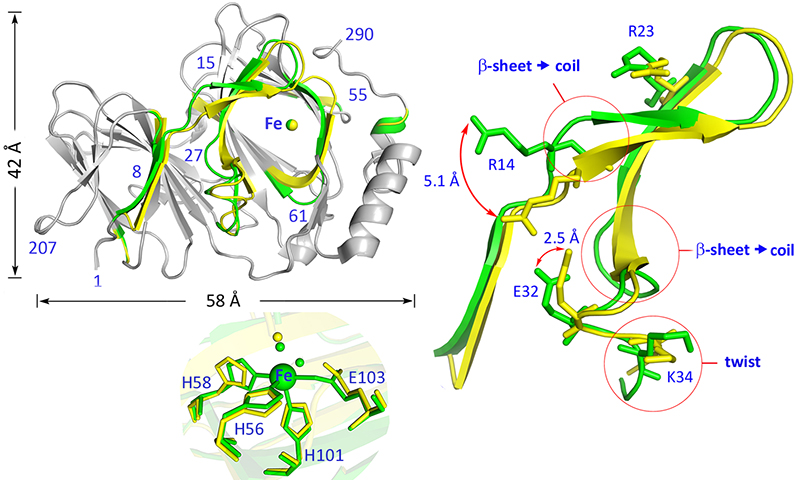
Pirin is a non-heme iron-dependent redox regulator of NF-κB
Liu F, Rehmani I, Esaki S, Fu R, Chen L, Serroano V, and Liu A*
PNAS 2013, 110(24), 9722-9724 (DOI: 10.1073/pnas.1221743110).
Structures associated with this publication and released in the protein databank with the following PDB entries: 4GUL, 4EWA, 4EWE, 4ERO, 4EWD, and 4HLT.
(This paper reports our discovery of a novel non-heme iron protein-based redox sensor in cell nucleus for NF-κB signal transduction regulation).
The Power of two: arginine 51 and arginine 239* from a neighboring subunit are essential for catalysis in α-amino-β-carboxymuconate-ε-semialdehyde decarboxylase
Huo L, Davis I, Chen L, and Liu A*
J. Biol. Chem. 288, 30862-30871 (DOI: 10.1074/jbc.M113.496869
Structures associated with this publication and released in the protein databank with the following PDB entries: 4IFK, 4IFO, 4IFR, and 4IG2.
In this work, we solved a catalytically active protein structure from a heterogeneous solution of simple mix of two dead mutant homodimers! In the resulting hybrid heterodimer one subunit
contains a wild-type structure while in another subunit a double mutation is present. Such a catalytically active hybrid cannot be
generated by molecular biology methods!.
Human α-amino-β-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD): A structural and mechanistic unveiling
Huo L,^ Liu F,^ Hiroaki I, Li T, Hasegawa Y, and Liu A*
PROTEINS 2014, 83(1), 178-187 (featured as a cover story by the journal).
Structures associated with this publication and released in the protein databank with the following PDB entries:: 4OFC, 4IH3, and 4IGN.
^: shared first authorship
This paper reports the crystal structure of catalytically active form of human ACMSD enzyme and the complex structure with bound ligand.
Evidence for a dual role of an active site histidine in α-amino-β-carboxymuconate-ε-semialdehyde decarboxylase
Huo L et al.
Biochemistry 51, 5811-5821.
Structures associated with this publication and released in the protein databank with the following PDB entries: 4ERA, 4ERG, 4ERI, and 4ERK
This paper reports a single mutation at the second ligand sphere (non-metal ligand) changes a decarboxylase's metal selectivity from zinc to iron.
Twenty other structures deposited and ready for publication in near future
Ruturn to Homepage